How To Run Logistic Regression In R
For this post I will use Weely stock market S&P data between year 1990 and 2010. I downloaded the data from following link...
app.quadstat.net/dataset/r-dataset-package-islr-weekly
How to read csv data in R
df = read.csv('data/dataset-95529.csv',header = TRUE)
Let us check the number of rows in our R dataframe using nrow.
nrow(df)
For columns, we can use ncol(dataframe)
ncol(df)
Data has 9 columns. All the columns are self explanatory except lag1,lag2,lag3,lag4,lag5 which are percentage returns for previous weeks.
Let us look at the summary of our data. We can use summary function in R which takes the dataframe and prints valuable summary.
summary(df)
In our summary above, we can see that last column is "Direction". Out of 1089 entries, 484 times it tells us that market had negative return and 605 times positive return.
We can use this data to train our model to predict if the weekly return would be positive or negative.
How to run Logistic Regression in R
Since the variable "Direction" is categorical. We can try using Logistic Regression. Logistic regression is similar in nature to linear regression. In R it is very easy to run Logistic Regression using glm package. glm stands for generalized linear models. In R glm, there are different types of regression available. For logistic regression, we would chose family=binomial as shown below.
glm.fit <- glm(Direction ~ Lag1 + Lag2 + Lag3 + Lag4 + Lag5 + Volume, family = binomial, data = df)
glm.fit is our model. glm is the package name. Direction is the output variable. To right of symbol ~ everything else is independent variables.
We can look at the summary of our logistic model using function summary.
summary(glm.fit)
summary has lot of information. We can selectively look at the information too. To check what are the available fields to query in the summary, do names(summary(model)).
names(summary(glm.fit))
Let us save the summary in to new variable and then query some of the above fields.
glm.sum <- summary(glm.fit)
Let us query coeffients of our logistic regression model.
glm.sum$coefficients
Above matrix is very important. The last column Pr(>|z|) is a p-value. If Pr(>|z|) is less than 0.05, it means parameter is significant and tells us that co-efficient estimate is significantly different from zero. All the parameters which have Pr(>|z|) less than 0.05 are significant. In the above table, we can see that intercept, Lag2 have p-value less than 0.05, there are significant parameters.
Let us use our model now to predict. In practice we should train our model on training data, then test it on unseen data. For now we are skipping that part. We would take our previous model which has already seen our test data.
glm.probs = predict(glm.fit,type="response")
Ok, our predict model is ready. Remember this is logistic regression, so our model would generate probabilities. We would mark our return as Up if probability is greater than 0.5 otherwise down.
glm.pred = rep("Down",length(glm.probs))
glm.pred[glm.probs > 0.5] = "Up"
Let us now look at the output in the form of confusion matrix.
table(glm.pred, df$Direction)
the above confusion matrix: Error rate (Down) = 430/(430+54) = 88.8% that means 88.8% of predictions about the down days are wrong , for all these days the model has predicted that market will go up. Error rate (Up) = 48/(48+557) = 7.9%, whereas while predicting Up days, the model has done very good job of being wrong only 7.9%
How to run Logistic Regression in R using Deep Learning library H2o
We can improve on our previous Logistic Regression results using deep learning package from H2o library.
Make sure you have h2o installed. If not Check out following tutorial to install h2o.
Once you have h2o installed. Let us import h2o and initialize it.
library(h2o)
h2o.init()
Let us first import our data using h2o.importFile function.
df.h2o <- h2o.importFile('data/dataset-95529.csv')
Let us define a variable to store all the x variables. We would use -match function in R to do that.
xall <- names(df.h2o)[-match(c("Direction"),names(df.h2o))]
head(df.h2o,1)
head(df[xall],1)
This time around, we would test our model against unseen data. Let us split the data in to train, valid and test data using h2o.splitFrame function in R as shown below.
parts <- h2o.splitFrame(df.h2o,c(0.8,0.1),seed=70)
train <- parts[[1]]
valid <- parts[[2]]
test <- parts[[3]]
xall
Let us now build our h2o deeplearning model. We would wrap it around with system.time to see the time taken to build the model.
y <- 'Direction'
system.time(m <- h2o.deeplearning(xall,y,train,validation_frame = valid))
Ok, model building was pretty quick. Let us look at the perfomance on validation set.
h2o.performance(m,valid = TRUE)
Fron the confustion matrix, we can see model is spot on. Model has been able to predict everything right.
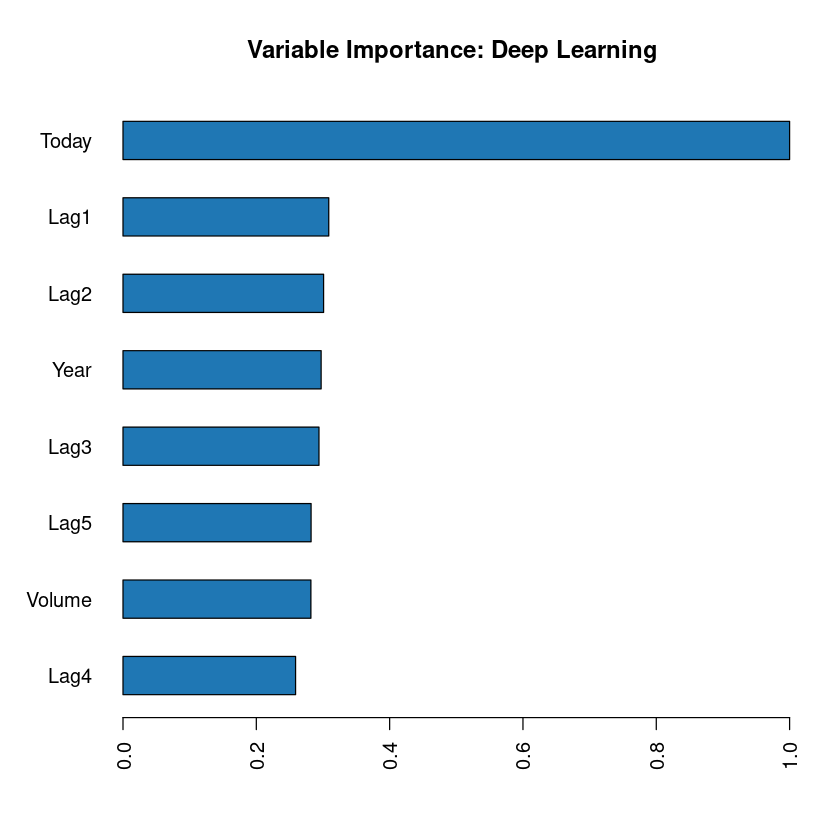
To see the importance of each variable in our model. We can use h2o.varimp_plot() function.
h2o.varimp_plot(m)
As see see above, variable "Today" (price) is the most important one, followed by Lag1 and so on and soforth.
Let us see now how our model performs work the unseen data. We would feed in test data which is not seen by our model so far yet.
h2o.performance(m,test)
Ok, our model has done pretty well. Predicting everything correct. We can also look at our confusion matrix using h2o.confusionMatrix as shown below.
h2o.confusionMatrix(m,test)
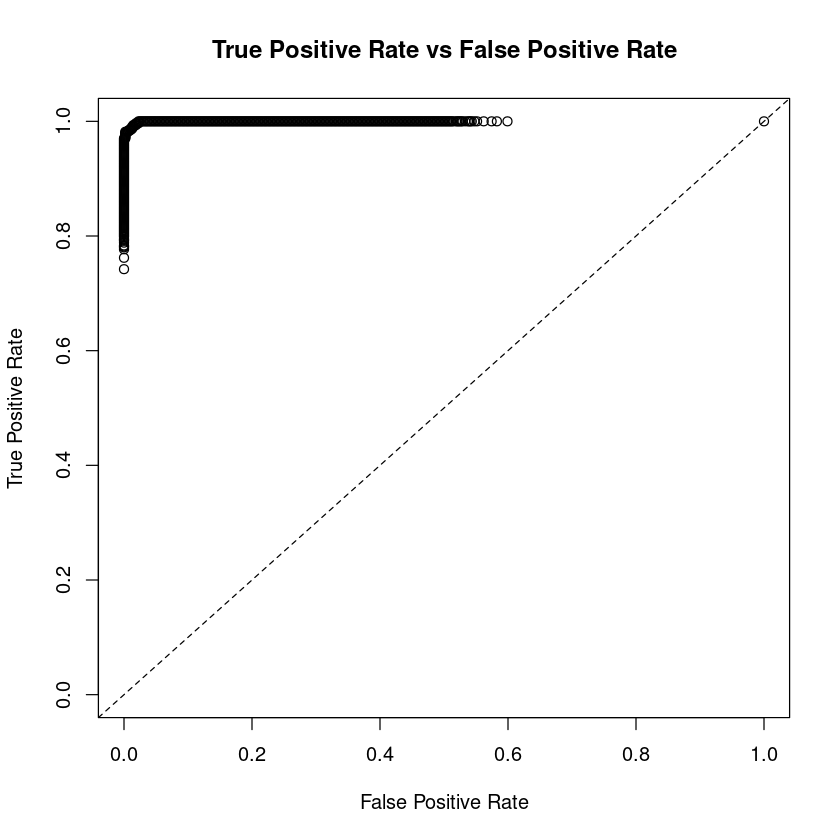
Let us end this post by plotting ROC curves. ROC curves plot "True Positive Rate" vs "Fals Positive Rate".
- True Positive Rate (Sensitivity) - The probability of target = Y when its true value is Y
- False Positive Rate (Specificity) - The probability of Target = Y when its true value is not Y
Ideally the ratio between ROC curve and diagonal line should be as big as possible which is what we got in our model. The plot is shown below.
perf <- h2o.performance(m, df.h2o)
plot(perf, type="roc")
Wrap Up!
In this post, I have demonstrated how to run logistic regression with R. Also exposed you to machine learning library H2o and its usage for running deep learning models.Related Notebooks
- Understanding Logistic Regression Using Python
- How To Run Code From Git Repo In Collab GPU Notebook
- How To Add Regression Line On Ggplot
- Regularization Techniques in Linear Regression With Python
- Decision Tree Regression With Hyper Parameter Tuning In Python
- How To Write DataFrame To CSV In R
- How To Plot Histogram In R
- How To Use Grep In R
- How To Iterate Over Rows In A Dataframe In Pandas